Intro
 GLinker is a modular entity linking framework that transforms raw text into structured, disambiguated entity mentions. It combines Named Entity Recognition, multi-layer database search, and neural entity disambiguation into a unified, configurable pipeline. GLinker is built around GLiNER — a family of lightweight, generalist models for information extraction — bringing zero-shot recognition, efficient BiEncoder support, and a compact inference stack to the entity linking task.
GLinker is a modular entity linking framework that transforms raw text into structured, disambiguated entity mentions. It combines Named Entity Recognition, multi-layer database search, and neural entity disambiguation into a unified, configurable pipeline. GLinker is built around GLiNER — a family of lightweight, generalist models for information extraction — bringing zero-shot recognition, efficient BiEncoder support, and a compact inference stack to the entity linking task.
What is Entity Linking?
Entity linking (EL) is the task of connecting textual mentions of entities to their corresponding entries in a knowledge base (e.g., Wikidata, custom databases). Unlike NER, which only detects entity spans and types, entity linking resolves ambiguity by determining which specific entity a mention refers to.
For example, the mention "Apple" in a text could refer to Apple Inc. (the technology company), apple (the fruit), or Apple Records (the record label). Entity linking disambiguates the mention and connects it to the correct knowledge base entry.
Traditional vs GLinker Approach
# Traditional approach: Complex, coupled code
ner_results = spacy_model(text)
candidates = search_database(ner_results)
linked = gliner_model.disambiguate(candidates)
# Mix of models, databases, and business logic
# GLinker approach: Declarative configuration
from glinker import ConfigBuilder, DAGExecutor
builder = ConfigBuilder(name="biomedical_el")
builder.l1.gliner(model="knowledgator/gliner-bi-base-v2.0", labels=["gene", "protein", "disease"])
builder.l2.add("redis", priority=2).add("postgres", priority=0)
builder.l3.configure(model="knowledgator/gliner-linker-large-v1.0")
executor = DAGExecutor(builder.get_config())
result = executor.execute({"texts": ["CRISPR-Cas9 enables precise gene therapy"]})
Architecture
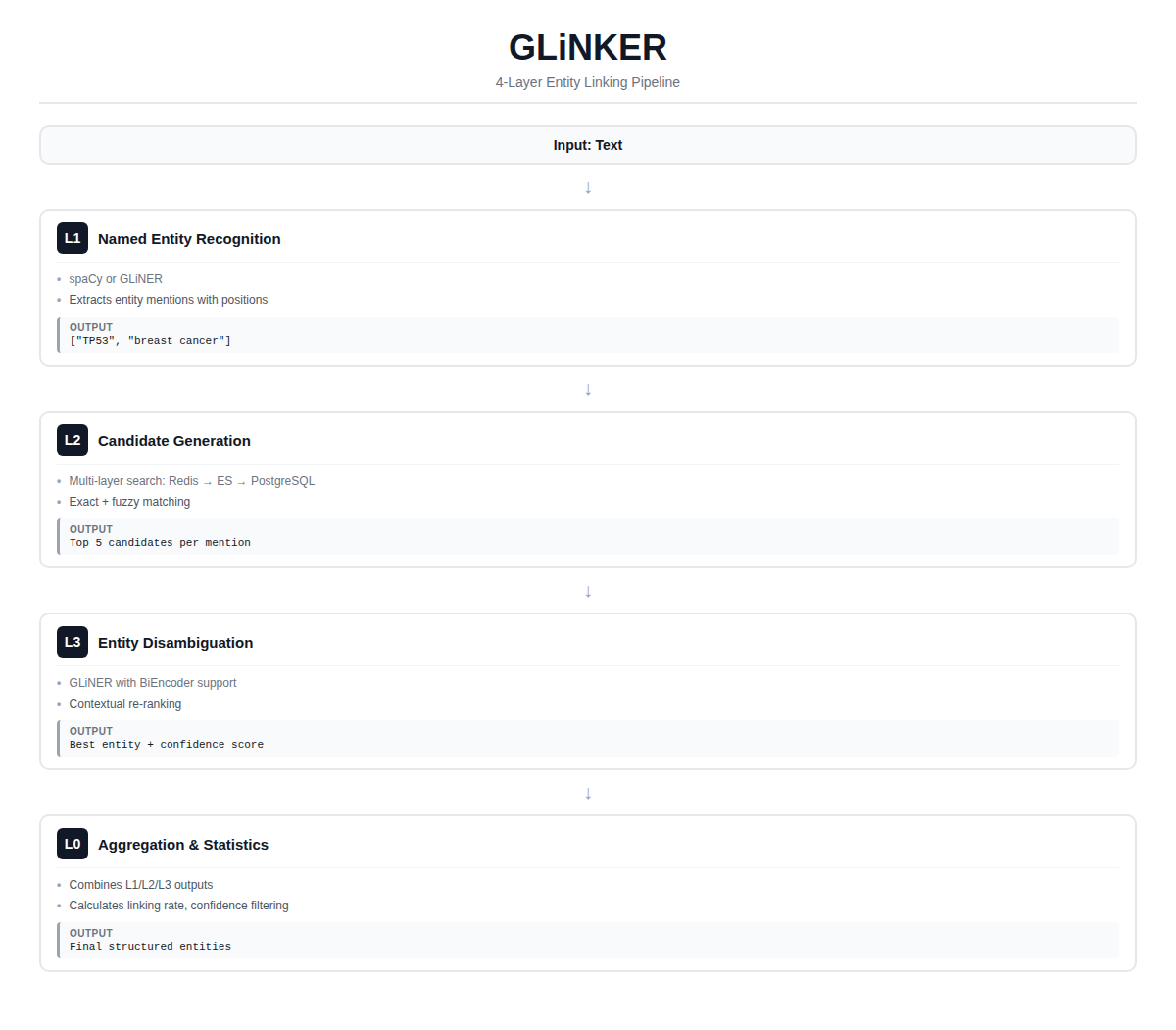
GLinker uses a layered pipeline with an optional reranking stage:
| Layer | Purpose | Processor |
|---|---|---|
| L1 | Mention extraction (spaCy or GLiNER NER) | l1_spacy, l1_gliner |
| L2 | Candidate retrieval from database layers | l2_chain |
| L3 | Entity disambiguation via GLiNER | l3_batch |
| L4 | (Optional) GLiNER reranking with candidate chunking | l4_reranker |
| L0 | Aggregation, filtering, and final output | l0_aggregator |
Supported Topologies
Full pipeline: L1 → L2 → L3 → L0
With reranking: L1 → L2 → L3 → L4 → L0
Simple (no NER): L2 → L3 → L0
Simple + reranker: L2 → L4 → L0
L1: Named Entity Recognition
The NER layer detects entity mentions in the input text. GLinker supports multiple NER backends:
- GLiNER models — Zero-shot NER with support for arbitrary entity types. Identify any entity type by simply providing label names, no fine-tuning required.
- spaCy models — Traditional NER with pre-defined entity types. Fast and rule-based for standard use cases.
L2: Candidate Retrieval
The candidate retrieval layer searches one or more entity databases to find potential matches for each detected mention. Supported backends:
- Dict — In-memory dictionary, perfect for demos and small-scale use
- Redis — High-performance key-value store for fast caching
- Elasticsearch — Full-text search with fuzzy matching for large-scale deployments
- PostgreSQL — Persistent storage with pg_trgm fuzzy search
L3: Entity Disambiguation
The disambiguation layer uses GLiNER-based linker models to rank candidate entities and select the best match for each mention. It computes similarity between mention context and entity descriptions.
L4: Reranking (Optional)
When the candidate set from L2 is large (tens or hundreds of entities), a single GLiNER call may be impractical. The L4 reranker solves this by splitting candidates into chunks:
100 candidates, max_labels=20 → 5 GLiNER inference calls
Results merged, deduplicated, filtered by threshold
L0: Aggregation
The aggregation layer combines results from all previous layers into a final, consistent output format with filtering and confidence scoring.
Key Concepts
DAG Execution
GLinker pipelines are structured as Directed Acyclic Graphs (DAGs). Each layer can depend on outputs from previous layers, enabling both simple linear pipelines and complex branching configurations. Layers execute in dependency order with automatic data flow.
Component-Processor Pattern
Each pipeline layer follows a component-processor pattern:
- Components handle the core methods (model inference, database queries)
- Processors orchestrate data flow, pre/post-processing, and batching
Schema Consistency
A single template (e.g., "{label}: {description}") is shared across layers, ensuring that entity representations remain consistent throughout the pipeline.
Cache Hierarchy
GLinker implements multi-level caching for production use:
- Upper database layers (Redis) cache results from lower layers (PostgreSQL) automatically
- BiEncoder models support embedding precomputation for 10-100x speedups
- On-the-fly embedding caching during inference
Why GLiNER for Entity Linking?
GLinker is built around GLiNER, which brings several key advantages:
- Zero-shot recognition — Identify any entity type by providing label names. Switch from biomedical genes to legal entities by changing a list of strings.
- Unified architecture — A single model handles both NER (L1) and entity disambiguation (L3/L4), reducing deployment complexity.
- Efficient BiEncoder support — Pre-compute label embeddings once and reuse them across millions of documents.
- Compact and fast — Base models run on CPU, while larger variants scale with GPU for production throughput.
- Open and extensible — Apache 2.0 licensed models on Hugging Face, easy to swap for domain-specific fine-tunes.
Use Cases
Biomedical Text Mining
builder.l1.gliner(
model="knowledgator/gliner-bi-base-v2.0",
labels=["gene", "protein", "disease", "drug", "chemical"]
)
News Article Analysis
builder.l1.spacy(model="en_core_web_lg")
# Link to Wikidata/Wikipedia entities
Clinical NLP
builder.l1.gliner(
model="knowledgator/gliner-bi-base-v2.0",
labels=["symptom", "diagnosis", "medication", "procedure"]
)